| FBB2 | |
| Project ID | 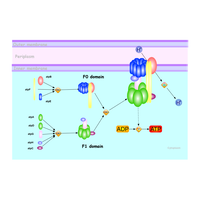 FBB2 [Protein] |
| Project Theme | Solid-state NMR investigation on functional and irregular structures of H+-ATPsynthase Fo |
| Project Theme (short) | H+-ATPaseF0 |
| Principal Investigator | Hideo Akutsu |
| Affiliation | Institute for Protein Research, Osaka University |
| Backgrounds | - ATP is the common energy currency of cells - H+-ATP synthase (F1-FO ATP synthase) that can synthesize ATP is of crucial importance in almost all organisms - The FO portion is within the membrane and its structure is unknown - We will solve the structure of the ring of c-subunits of FO of bacterial ATP synthase using solid-state NMR |
| Highlights | - Using membrane-reconstituted subunit c-ring of E.coli H+-ATP synthase, we have shown that the high energy conversion efficiency of the synthase is due to the smooth rotation of the c-ring - A subunit packing model of E. coli c-ring has been proposed - The main chain secondary structure of thermophile c-ring has been obtained |
| Outline | ATP synthase is a general term for an enzyme that can synthesize adenosine triphosphate (ATP) from adenosine diphosphate (ADP) and inorganic phosphate by using a form of energy. These enzymes are of crucial importance in almost all organisms, because ATP is the common energy currency of cells. H+-ATP synthase (F1-FO ATP synthase) is a large complex consisting of numerous subunits and is divided into Fo and F1 particles. The Fo portion is within the membrane and the F1 portion looks like a propeller with 6 blades inside the membrane. A portion of the FO (the ring of c-subunits) rotates as the protons pass through the membrane. The c-ring is tightly attached to the central stalk which rotates the axis of F1 causing the 3 catalytic nucleotide binding sites to go through a series of conformational changes that leads to ATP synthesis. We will solve the structure of the ring of c-subunits of FO of bacterial ATP synthase using solid-state NMR. |
| Review | - |
| CSML File | FBB2.csml |